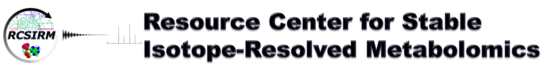You are here: Moseley Bioinformatics Lab>RCSIRM Web>ServicesAndRates (27 Feb 2019, HunterMoseley)
Services and Rates
Levels of Service
The Center has several levels of interaction with Clients as follows:
- Basic Analytical Services
- The Client will supply raw tissue/cells/media with a request for sample processing with NMR and/or MS analyses
- OR the Clients supply metabolite extracts with a request for standard NMR and/or MS analyses.
- Then the Center prepares the extracts accordingly, records the appropriate spectra under standard operating conditions, and returns the raw data to the Client
- OR the Center prepares the extracts accordingly, records the appropriate spectra under standard operating conditions, and reduces the data to a list of quantified identified metabolites with respect to the current databases, with isotopomer and isotopologue distributions when appropriate.
- Experimental Design
- The Client consults with the Center leadership on the overall goals to develop a detailed plan for the experimental design, numbers of samples, sample handling and integrity, numbers of analytes and analytical platforms, number and types of tracers to be used.
- Biostatistical Analysis
- The Center will assist in various biostatistical analyses including metabolic profiling and sample classification.
- Guidance on Mechanism-Based Analysis and Omics Integration
- The Center will assist in mechanism-based biochemical interpretation, which includes relevant metabolic pathway analysis and integration of metabolic data with other streams of information such as transcriptomics, proteomics, and functional annotations.
- Beyond rudimentary guidance, this will require collaboration.
- Custom Services
- The Center will work with Clients to develop new methods and standard operating procedures that address their specific needs.
- In vivo NMR experiments (e.g. preclinical mouse model studies) will always be under this category.
- Custom Services will usually incur the highest rates, and will require collaboration where new methods or assays are developed
Academic Rate Schedule
All rates stated are based on estimates from recharge rate calculation spreadsheets (RRC) that annually balance Center expenditures with projected income for each chargeable operation (e.g. each instrument, sample preparation technique, informatics computer usage).
2017-2018 Sample & Analytical Cores Rates for not-for-profit
* Excel Rates Table for download.*
Note that some rates are per run, some are per hour (smallest increment), and some are surcharges.
* commercial rates are 4-6 times higher than academic rates *
* rates are presently under review *
**available only to academic users, not to fee for service commercial users
| Cost Rate # | Title | Short Description | Description | Per Sample | Surcharge Per Run | Per Hour |
|---|---|---|---|---|---|---|
| 1 | Tier A FTMS | Non-polar, Comprehensive | Detailed, non-polar analysis, e.g. involving global by-lipid class analysis but with more exhaustive, custom investigations. Likely surcharges include Tiers G, H, I (Rates 7, 8, 9) and Data Processing (Rate 31). | $155.00 | ||
| 2 | Tier B FTMS | Non-polar, General/Typical | Typically performed e.g. lipids analysis involving by-class distributions, ratios and selected fractional enrichment. Likely to incur Data Processing (Rate 31) surcharges, especially for data-mining non-assigned spectral features. | $75.00 | ||
| 3 | Tier C FTMS | Non-polar, Limited/Targeted | Targeted analysis of a defined, short list of analytes, e.g. 10 specific isobaric lipids. Usually performed in a fashion that may preclude data mining for non-target analytes. [Note: Proximate type of analysis, such as total diacyl glycerides, is NOT this rate, but is rather Tier B (Rate 2).] | $45.00 | ||
| 4 | Tier D FTMS | Polar, Comprehensive | Full, detailed analysis of major polar compounds, e.g. a global nucleotide analysis with other more exhaustive, custom investigations. Likely surcharges include Tiers G, H, I (Rates 7, 8, 9) and Data Processing (Rate 31) | $120.00 | ||
| 5 | Tier E FTMS | Polar, General/Typical | Typically performed analysis involving polar metabolite distributions, ratios and selected fractional enrichment. Likely to incur Data Processing (Rate 31) surcharges, especially for data-mining non-assigned spectral features. | $75.00 | ||
| 6 | Tier F FTMS | Polar, Limited/Targeted | Targeted analysis of a defined, short list of analytes, e.g. 10 specific nucleotides. Usually performed in a fashion that may preclude data mining for non-target analytes. [Note: Proximate type of analysis, such as total purine nucleotides, is NOT this rate, but is rather Tier E (Rate 5).] | $45.00 | ||
| 7 | Tier G FTMS | Multiply labeled tracer analysis (for example 15N13C Gln) | Analysis is complicated and time consuming as more than one stable isotope enrichment is investigated. Likely to incur Data Processing (Rate 31) surcharges. | $45.00 | ||
| 8 | Tier H FTMS | Tandem MS analysis | Manual/Non-automated analysis of MS/MS spectra for structural confirmation. Likely to incur Data Processing (Rate 31) surcharges. | $45.00 | ||
| 9 | Tier I FTMS | Unknown structural assignment | Comprises of multiple approaches - time consuming and non-automatic. Likely to incur Data Processing (Rate 31) surcharges. | $75.00 | ||
| 10 | Tier J FTMS | MS1 only, no data analysis | Acquisition of FTMS spectrum, no data analysis. | $30.00 | ||
| 11 | Tier K LC use | LC use | LC use surcharge. LC includes ion chromatography, now used extensively for anionic metabolites | $60.00 | ||
| 12 | Tier L GCMS | Comprehensive, In-depth analysis | Full, detailed, involving not only a global analysis but also other more exhaustive, custom investigations inluding, but not limited to: unknowns, etc. Likely surcharges include Tiers G, H, I (Rates 7, 8, 9) and Data Processing (Rate 31). | $140.00 | ||
| 13 | Tier M GCMS | General/Typical | Typically performed analysis involving by-class distributions, ratios and selected fractional enrichment. Likely to incur Data Processing (Rate 31) surcharges, especially for data-mining non-assigned spectral features. | $55.00 | ||
| 14 | Tier N GCMS | Highly Targeted or Data Only | Usually a targeted analysis towards a defined, short list of analytes, e.g. a drug and its major metabolite. Usually performed in a fashion that may preclude data mining for non-target analytes. | $27.00 | ||
| 15 | Specimen Grinding | Specimen Grinding (e.g. LN2 grinding) | Tissue grinding & cryo-grinding. | $20.00 | ||
| 16 | Specimen Polar Extraction | Specimen Polar Extraction | Polar metabolites extraction. | $15.00 | ||
| 17 | Specimen Non Polar Extraction | Specimen Non Polar Extraction | Non-Polar metabolites extraction. | $15.00 | ||
| 18 | Sample Lyophilization | Lyophilization operations | Lyophilization. | $10.00 | ||
| 19 | Ultracentrifugation | Ultracentrifuge processing | Ultracentrifugation up to 120K xg. | $15.00 | ||
| 20 | Digester/Synthesizer | CEM Explorer Focused Microwave | Acid digestions or microwave synthesis runs. | $25.00 | ||
| 21 | 14.1T NMR | 14.1T NMR, RCSIRM-provided operator and Data Only | All 1D and multidimensional NMR experiments are covered in this hourly rate, run by a RCSIRM operator. | $25.00 | ||
| 22 | 14.1T NMR User | 14.1T NMR, Trained user** | All 1D and multidimensional NMR experiments are covered in this hourly rate for a user that has completed training. | $12.00 | ||
| 23 | 16.45 T NMR user | RCSIRM operator | All 1D and multidimensional NMR experiments are covered in this hourly rate, run by a RCSIRM operator. | tbd | ||
| 24 | 16.45 NMR Trained User | Data Only | ||||
| 25 | 9.4T NMR Widebord | Small Animal: per hr/4 hr min | $44.00 | |||
| 26 | ICPMS | QqQ -ICPMS | Single and multi-element, with & without reaction cell. | $90.00 | ||
| 27 | Protein Determination | Total protein determination | For many types of samples, it is best to normalize analyte results to total protein. | $20.00 | ||
| 28 | Robotic Liquid Handler | Robotic Liquid Handler | Standalone use of the Tecan Fluent 780 liquid handler | $40.00 | ||
| 29 | Particle Analyzer | Nanosight Particle Counter/Sizer | Counts particles with size distribution, e.g. microvesicles. | $10.00 | ||
| 30 | Tubewriter | Tubewriter labeler | Automated labware labeler capable of 1 & 2D barcodes. | $10.00 | ||
| 31 | Data Processing | Data Processing | Data processing where not included above, or as a surcharge. | $40.00 | ||
| 32 | Ancillary Labor | Ancillary Labor | Method development in excess of rates 1-31, extensive advising and instruction | $60.00 | ||
| 33 | Ancillary Supplies | Ancillary Supplies | Method development in excess of rates 1-31, extensive advising and instruction | at cost | ||
| 34 | Bench Fee | Half Bench for sample processing, 11 linear ft per day | For clients lacking sufficient facilities may utilize RCSIRM wet bench. User must meet all regulations and requirements for lab bench work at UKY, including any specific requirements for working in RCSIRM space (which handles e.g. human subject samples). | $66.00 |
Supplies
Common Isotopically Enriched Precursors
| Compound | CIL # | Isotec # |
|---|---|---|
| [U-13C]-glucose | CLM-1396-PK * | 389374-1G * |
| [U13C, 15N]-Gln | CNLM-1275-H-PK | 607983-500MG |
| 13C1,2 Glucose | CLM-504-1 * | 453188-1G |
| [U-13C, 15N] L Serine | CNLM-474-H-0.5 | 608130-250MG |
| [U-13C]-octanoate | CLM-3981-PK | 605727 |
| [U-13C]-palmitate | CLM-409-0.5 | 605573-100MG |
| 99.9% D2O | DLM-11-100 | 151882-100G |
| 70% D2O | DLM-2259-70-1L | 613428-1KG |
- * means not microbiology nor S*&P tested. OK for cell culture.
- Conversions:
- 1 g glucose = 5.37 mmol
- 0.5 g Gln = 3.27 mmol
- 0.5 g serine = 4.13 mmol
- 0.5 g palmitate = 1.85 mmol
NMR Tubes
- 3 mm Shigemi: $95 each
- Wilmad Tubes
- Regular 3 mm (Wilmad): $17.18 each - 335-PP-7
- Regular 5 mm (Wilmad): $19.19 each - 535-PP-7
Informatics Core
All non-automated and semi-automated data analyses will cost $50/h for academic Clients and $150/h for industrial Clients.
- All automated analyses, requiring less than 1 CPU-core h of execution time, will cost $5 per sample dataset.
- Automated analyses requiring more than 1 CPU-core h will be charged $5 for the first CPU-core h and $1 for each additional CPU-core h.
Service Examples
13C Dispersal from Glucose into Krebs Cycle, Glycolysis
| Cost Rate | Rate/Sample |
|---|---|
| Sample Processing | $30 |
| Tier M GCMS | $55 |
| 14.1 T NMR | $20a |
| Total= | $117 |
Thus per sample charges are estimated.
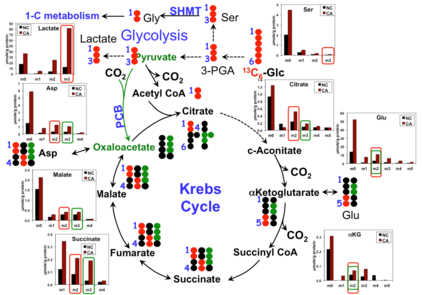
13C and 15N Dispersal from Glutamine into Mononucleotides
| Cost Rate | Rate/Sample |
|---|---|
| Sample Processing | - |
| Tier E FTMS | $75 |
| 14.1 T NMR | $25a |
| Total= | $140b |
Thus per sample charges are estimated.
bFurther detailed isotope labeling is data processing costs only.
"Global" 13C Lipidomics from labeled Glucose or Glutamine
| Cost Rate | Rate/Sample |
|---|---|
| Sample Processing | $30 |
| Tier B FTMS | $75 |
| Additional Data Processing | $40a |
| Total= | $145a |
depending on level of detail required.
Key Questions
Before contacting the Center for services, researchers should be prepared to discuss the following:
- What is the scientific question or problem?
- Examples:
- I need to determine the change in molecule X when I do Y.
- I want to know whether the oxidative branch of the pentose phosphate pathway is contributing to the production of NADPH.
- Examples:
- What is the experimental system?
- Cell culture, spheroids, animal models, human subject, PDX, etc.
- Targeted or global metabolomics? Pathway analysis or flux analysis?
- For these questions, often the researcher will not have an answer, but should expect that these aspects will be discussed.
- It is highly recommended that the client be sure of the term flux, and refresh themselves on network dynamics and reaction diffusion systems under inhomogeneous conditions.
- What information is already available?
- Transcriptomics data may imply metabolic consequences and may be partially validated by protein analysis (cf. Westerns, MS, enzyme activity).
- The Seahorse XRF provides information about the rate of proton extrusion (ECAR) and the rate of oxygen consumption (OCR) by adherent cells under normoxic conditions.
- By varying the extracellular nutrient source (e.g. glucose, glutamine, fatty acids) and with the use of specific inhibitors, it is possible to estimate net lactic fermentative flux, glycolytic reserve, respiration potential and the degree of coupling to oxidative phosphorylation. Such information is extremely valuable in designing metabolomics analyses or whether metabolism is a significant correlate of the biological function of interest.
- The consultation will then focus on the experimental design, which will also need to include biostatisticians for estimating sample sizes.
- It is also important to establish whether the user will do their own sample work up, what analytical techniques are needed, and what level of data reduction is required.
- The Center has SOPs. Questions that cannot be answered using these SOPs will be considered in terms of development time to establish specific metabolic assays.
- This is relevant mainly in the context of a small number of metabolites (targeted metabolomics).
- Costs.
- What are the costs associated with the samples being analyzed?
- What are the costs of the metabolomics experiments, including tracers?
- It is VERY common for data analysis to take 90% of the metabolomics labor, so the researcher should be prepared to discuss the time frame and cost structure with this consideration, versus obtaining data analysis training for their own laboratory.
- We have found, however, that the latter approach still incurs considerable labor for the RCSIRM staff, as data analysis is highly nuanced and extensive consulting is still typically needed.
-
Once an experimental approach has been worked out, the user will be introduced to laboratory personnel who will carry out the analyses and oversee the project progress- this person will then be the primary contact.
Data collection is not the rate limiting step. Global analyses often produce hundreds to thousands of identifiable and quantifiable compounds or features, but this means the reduction of raw data is extremely time consuming.
Useful Publications
Examples of the use of stable isotope resolved metabolomics can be found in the publications listed below:
-
- Fan, T.WM., Lane, A.N., Higashi, R.M., Farag, M.A., Gao, H., Bousamra, M. & Miller, D.M. (2009) Altered Regulation of Metabolic Pathways in Human Lung Cancer Discerned by 13C Stable Isotope-Resolved Metabolomics (SIRM). Molecular Cancer. 8:41
- Fan, T. W-M., Lane, A.N., Higashi, R.M., Yan, J. (2011) Stable Isotope Resolved Metabolomics of Lung Cancer in a SCID Mouse Model. Metabolomics 7, 257-269
- Le, A., Lane, A.N., Hamaker, M., Bose, S., Barbi, J., Tsukamoto, T., Rojas, C.J., Slusher, B.S., Zhang, H., Zimmerman. L.J., Liebler, D.C., Slebos, R.J.C., Lorkiewicz, P.K., Higashi, R.M., Fan, T.W-M., and Dang, C.V. (2012) Myc induction of hypoxic glutamine metabolism and a glucose-independent TCA cycle in human B lymphocytes. Cell Metabolism. 15, 110-121
- Yang, Y., Lane, A.N., Fan, T.W-M., Rickets, C., Wu, M., Boros, L., Linehan, W.M. (2013) Understanding How Fumarate Hydratase (FH) Null Cells Use its Central Carbon for Energy and Malignant Development PlosOne 8, e72179
- Sellers, K., Fox, M.P., Bousamra, M., Slone, S., Higashi, R.M.,Miller, D.M., Wang, Y., Yan, J., Yuneva, M., Deshpande, R., Lane, A.N., Fan, T. W-M. (2015) Pyruvate carboxylase is upregulated in NSCLC. J Clin Invest. 125(2): 687-698
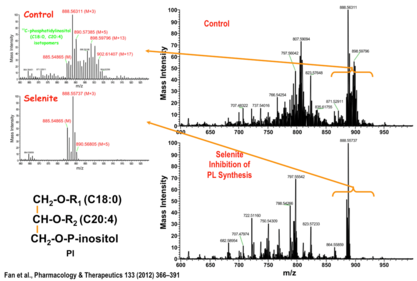
- The review by Fan, T.W-M., Lorkiewicz, P., Sellers, K., Moseley, H.N.B., Higashi, R.M., Lane, A.N. (2012). Stable isotope-resolved metabolomics and applications to drug development. Pharmacology & Therapeutics. 133:366-391 is also a good background source of examples.
- Our Handbook of Metabolomics (The Handbook of Metabolomics Methods in Pharmacology and Toxicology, vol. 17 Humana Press 2012. DOI 10.1007/978-1-61779-618-0_4)
An example study and pipeline
What is the role of tumorin in tumor development and survival in TNBC?
The goal is to understand how central metabolism is impacted by the knockdown, including sources of NADPH needed for proliferation. The user may already have information from Seahorse analysis, and phenotypic effects of the knockdown. Cell cycle distribution analysis is also important for the overall interpretation.
(i) 3 cell lines ±shRNA against tumorin- triplicate experiments, 3 tracers ([U-13C]-Glc, [13C1,2]-Glc, [U-13C,15N]-Gln = 54 experiments.
Polar + non polar metabolites = 108 analytical samples.
Protein may be used for additional experiments (e.g. expression), and/or normalization.
Polar metabolites analyzed by NMR and GC-MS or IC/FT-MS= 216 experiments,
+ 54 FT-MS of nonpolar fraction
total = 270 analyses.
Media samples at 5 time points for each dish =270 media samples, analyzed by NMR and MS
total= 540 analyses.
Total analyses = 810
(ii) Same cell lines as orthotopic xenograft in NSG mice (5 mice/group), two tracers = 60 mice.
Tumor tissue + nontumor tissue + 2 blood samples per mouse = 240 samples.
120 blood analyses by MS and NMR = 240 analytical samples.
Tissue polar and non polar = 480 analyses.
Total mouse sample analyses = 720
Grand total = 1530 analyses.
The number of analytes including isotopomers and isotopologues is > 200,000 quantified analytes for this study.
At this point, the data are reduced to lists of compounds, their isotopomers and their amounts for biological interpretation.
Outline Procedure
Design experiment->execute biological experiment with tracers-> harvest sample->prepare sample for analysis->analytical data acquisition->data reduction->information retrieval and interpretation.
Detailed SOPs are available
Cell culture
Grow cells in culture in triplicate with each tracer ± shRNA. 18 experiments (x3 for each cell line).
Sample media at defined time points and store (e.g. 0, 3,6,9,24 h-the zero time point is critical) (90 samples)
Harvest and extract cells
Store metabolite fractions (polar, non-polar, protein)
Dried samples must be reconstituted in appropriate volume of buffers for different analytical platforms, and loaded into the correct labeled sample tubes.
Prepare for NMR run first for quality control on sample and extraction integrity
Prepare for GC or IC MS
Prepare for FT-MS
Record spectra on the different platforms
Reduce data to raw isotopologues distributions for each tracer
Repeat any bad experiments
Repeat for next cell line
Tumor bearing mice:
treat with tracer, sample blood. Harvest tissue
Extract tissues and blood
Prepare for analytical spectroscopy
Record spectra on the different platforms
Reduce data to raw isotopologues distributions for each tracer
Repeat any bad experiments
Data acquisition and reduction.
GC-MS adn IC-MS take 1 h per sample to run. QC/standard samples must be run in interleaved mode.
NMR spectra take 1.2 h per cell or tissue sample to run, 0.5 h for plasma extracts
FT-MS for lipids takes 10-15 min per sample.
Data reduction for this density of data is 1-1.5 h/spectrum.
The results can be interpreted in terms of specific networks related to cell growth or survival, with limited flux information (exclusively in this design for inputs and outputs).
Data Processing and Analysis
The data comprise several components, as follows.
- Metadata that describe in exact detail the entire workflow from sample receipt to final products. No useful results can be obtained without these data. An Excel spreadsheet is required for these data, and a template is provided.
- Raw analytical data, i.e. the streams of bits coming from the instruments. For FT-MS and NMR these represent digitized electrical signals in the form of free induction decays comprising both real and imaginary parts. For other MS data, these are digital representations of ion counts.
- Raw analytical data have to be transformed into a usable form, which for FT-MS and NMR is the discrete fourier transform and associated digital processing to suppress truncation artefacts, optimize signal-to-noise ratios etc. The resulting output is a spectrum of intensity versus frequency. For NMR, the frequency is usually transformed to chemical shift, in ppm, that is independent of magnetic field strength. For FT-MS, the frequencies are mapped onto an m/z range.
- Intensities (ordinate values) must be internally normalized to obtain amounts of materials (i.e. numbers of moles of substances or of ions), and back calculated to the values associated with the original spectrum, on an agreed upon measure of the amount of that species (such as biomass weight, protein mass etc.). This absolutely requires accurate metadata. The amounts are proportional to peak areas (or volumes) NOT peak heights; appropriate numerical or analytical integration procedures must be correctly applied, taking due account of baseline drift, phasing errors and peak overlap.
- For isotopomer and isotopologue analyses, the intensities are usually expressed as mole fractions (enrichments). As these are ratios, normalization to cell amount is not needed. For MS, the natural abundance needs to be corrected. Software is available for this [cf. Moseley (2010) Correcting for the effects of natural abundance in stable isotope resolved metabolomics experiments involving ultra-high resolution mass spectrometry. BMC Bioinformatics 11,139]
- Spectral features need to be mapped onto identifiable molecules (assignment), using the available spectral information, and by reference to our own and other public databases.
- For profiling typically one is concerned with case-control comparisons, which require large numbers of specimens (each unique). Multivariate statistics are generally appropriate for initial analyses- are the groups different? What is different about these groups? PCA and OPLSDA (SimcaP) may be used.
- Normalization. To compare case and control, the quantity of each metabolite must be normalized to the appropriate amount of specimen. Cell number is generally not appropriate as cell volumes vary widely among types, and also in response to treatment. Total biomass or a surrogate is appropriate (e.g. dry weight, total protein total DNA).
- With SIRM studies, a question is often what pathways were impacted, which requires pathway tracing (SIRM) and biochemical expertise.
- With tracers, the time course of the isotopomer distributions can be determined, as can the fraction of glucose (glutamine) consumed that is converted to excreted product (e.g. lactate, alanine, glutamate).
- Further statistical analyses as needed should be carried out by statisticians versed in multivariate analyses.
Topic revision: r29 - 27 Feb 2019, HunterMoseley
 Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors. Ideas, requests, problems regarding Moseley Bioinformatics Lab? Send feedback