You are here: Moseley Bioinformatics Lab>RCSIRM Web>InformaticsCore (17 Jul 2016, AndrewLane)
Informatics Core
Overview
The Informatics Core provides necessary bioinformatic, biostatistical, and systems biochemical analysis tools as a set of automated and Client-guided services. These services have broad application from simple biomarker discover and profiling to mechanisms-based interpretation of biomarkers within the context of specific human disease processes. These services and tools are under continual development to keep up with improvements in experimental design and analytical capabilities. Currently, our development has a heavy emphasis on raw data analysis, metadata capture, and metabolite identification tools and services.
Web-based Informatics Platform
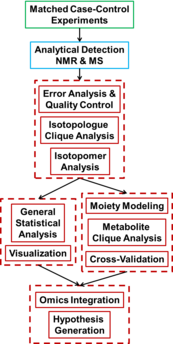 |
We are implementing a robust web-based data analysis pipeline that will facilitate all bioinformatics, biostatistical, and systems biochemical analyses used by the Center. The pipeline serves as a central point of metadata capture, which facilitates deposition of datasets into the Metabolomics Data Repository managed by Data Repository and Coordination Center. The pipeline also includes a wiki website that serves as a web-accessible documentation platform for all public documents and procedures of the Center. |
|---|
Platform Implementation
| Our platform implementation via an Apache web server using the Django web framework within a Linux virtual machine empowers independent development of underlying algorithms and tools, using a combination of languages and supporting libraries including Perl, Python, and R, while also enabling specific versioning of the whole data analysis pipeline. The TORQUE queuing system distributes calculations across a cluster of Linux computers. |  |
|---|
Raw Data Analysis Tools and Quality Control for MS and NMR Analytical Data
We are implementing standardized tools for analyzing FT-MS and NMR data with later expansion to GC-MS and LC/IC-MS data. These tools include:
- Peak assignment
- Metabolite identification
- Quantification and natural abundance correction
- Error analysis
- Quality control
- Identification and quantification of isotopologues and isotopomers
Basic Biostatistical Analyses
To promote sound interpretation of metabolomics data, we will provide some basic biostatistical analyses and visualizations for both SIRM and non-SIRM Client datasets. Furthermore, we promote the use matched case-control paired experimental designs, which allow the use of paired difference tests that have stronger statistical power. We will also provide a tiered set of biostatistical services that will use more advanced statistical and machine learning methods to detect sample separability and classify samples with respect to conditions and/or disease states. These services will evolve rapidly as we integrate more tools into the data analysis pipeline.
Additional tools are being developed and implemented for analyzing sparse high dimensional metabolomics data sets, and robust estimation of power.
(Eventually Coming) Metabolic Pathway Reconstruction, Flux Modeling, and Data Integration.
We will build upon our existing approaches that interpret stable isotope-resolved metabolomics (SIRM) data within the context of relevant metabolic networks and facilitate mechanism-based analysis of correlated metabolites of interest. Specifically, we are developing tools to automate reconstruction of metabolic networks that are relevant to the interpretation of SIRM time-series experiments and then determine the relative pathway fluxes through these networks. Furthermore, the resulting pathway-specific information serves as a point of integration with transcriptomic and proteomic data, allowing interpretation of specific biomarkers within the context of disease processes. These tools will provide the next level of hypothesis-generating and hypothesis-testing services that are instrumental to transformative discoveries in basic and translational research.
Other approaches to analyzing flux include: (i) functional fitting to time course data of media components (inputs and outputs) in cell culture such as glucose, glutamine, lactate, glutamate (ii) non-stationary state metabolic modeling of isotopomer/isotopolog distributions by numerical solutions to coupled differential equations representing a specific metabolic network (external collaborations).
Lane, A.N., Fan, T. W-M., Xie, X. Moseley, H.N. & Higashi, R.M. (2009) Stable isotope analysis of lipid biosynthesis by high resolution mass spectrometry and NMR. Anal.Chim. Acta. 651, 201-208 Moseley H.N.B. (2010) Correcting for the effects of natural abundance in stable isotope resolved metabolomics experiments involving ultra-high resolution mass spectrometry. BMC Bioinformatics 11,139 Moseley H.N.B. (2013) Error Analysis and Propagation in Metabolomics Data Analysis. Computational and Structural Biotechnology Journal. 4, e301006 Carreer ,W.J., Flight, R.M., Moseley, H.N.(2013) A Computational Framework for High-Throughput Isotopic Natural Abundance Correction of Omics-Level Ultra-High Resolution FT-MS Datasets. Metabolites 3, 853-866 Moseley, H.N.B., Lane, A.N., Belshoff, A.C, Higashi, R.M. Fan, W. W-M. (2011) Non-Steady State Modeling of UDP-GlcNAc Biosynthesis is Enabled by Stable Isotope Resolved Metabolomics (SIRM). BMC Biology 9, 37 Le, A., Lane, A.N., Hamaker, M., Bose, S., Barbi, J., Tsukamoto, T., Rojas, C.J., Slusher, B.S., Zhang, H., Zimmerman. L.J., Liebler, D.C., Slebos, R.J.C., Lorkiewicz, P.K., Higashi, R.M., Fan, T.W-M., and Dang, C.V. (2012) Myc induction of hypoxic glutamine metabolism and a glucose-independent TCA cycle in human B lymphocytes.Cell Metabolism. 15, 110-121 Mitchell, J.M., Fan, T. W-M., Lane, A.N., Moseley, H. N.B. Development and In silico Evaluation of Large-Scale Metabolite Identification Methods using Functional Group Detection for Metabolomics Frontiers in Genetics 5, 237
Topic revision: r7 - 17 Jul 2016, AndrewLane
 Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors. Ideas, requests, problems regarding Moseley Bioinformatics Lab? Send feedback