Sample Processing Core
Overview
Standard Sample Processing
- Subcellular fractions (e.g. exosomes),
- Cells
- Biofluids (plasma, urine, breath, nipple aspirate, etc.)
- Tissue
- Animal, plant, microbial and human tissues are extracted using appropriate protocols, e.g. for adherent cells with our rapid solvent quenching, followed by solvent partitioning methods to simultaneously obtain polar, non-polar, and protein fractions. This method is optimized for metabolite coverage (including relatively labile components), reproducible recovery, a wide range of sample sizes (µg to g weight), and compatibility with both NMR and MS.
- Initial sampling procedures, which is arguably the most critical step. In the NIH translational arena, the Core leads have hands-on experience and currently active research with human operating theater procedures, small animal and animal xenografts, GEMMs, human tissue slice experiments, and tissue cultures.
(Coming soon) Microfluidics-based subcellular/cell separation and metabolite extraction.
- High efficiency sorting of cells (e.g. leukocyte sub-populations including granulocytes, macrophages and lymphocytes) and capture of sub-cellular components (e.g. lipid microvesicles).
- High-speed phase partitioning of polar and non-polar metabolites.
- Ability to work with very small sample sizes.
Chemoselective Probes
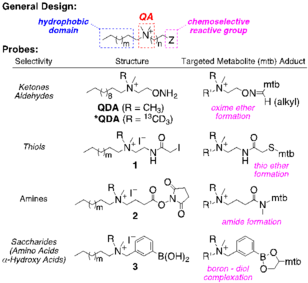 |
To capture low levels of polar carbonyl (C=O, ketone and aldehyde) metabolites directly from crude cell extracts, we have recently developed an aminooxy-based probe with quaternary ammonium (QA) and C12 alkyl functionalities (QDA). The resulting metabolite adducts are readily analyzed by infusion FT-ICR-MS at orders of magnitude lower levels than the parent metabolites, enabling further expansion of our analyte list by ≥300. We are actively extending CS tagging to metabolites containing amino (R-NH2), thiol (R-SH) and diol (R-CH(OH)CH(OH)-R) functionalities. The probes feature a modular design: selective reaction with the desired functional groups, a QA group for enhanced detection by [+] mode FT-ICR-MS, and a hydrophobic modifier group to facilitate sample cleanup. We are also synthesizing combinations of ≥10 isotope (13C, 15N, and 2H)-encoded CS probes for multiplexed analyses. Crude extracts of biospecimens can be treated with pairs of non-encoded and encoded CS probes, each pair having a CS tag targeting different functional groups, expanding on a strategy we have shown for tagging C=O groups in A549 cell extracts using QDA and 13CD3-QDA. FT-ICR-MS analysis quantifies the encoded adduct pairs resulting in the desired functional group profile(s). This sample encoding strategy will enable automated assignment software for metabolites and metabolically enriched isotopologues by the InformaticsCore, based on assigned functional groups and molecular formulae (MF), which is far more robust than MF alone. It will also enable tandem MS, LC-MS, and fast isotope-edited 2D NMR analysis to differentiate isomers and to discern substructures for de novo unknown assignment. The many isotope-encoded CS probes will also improve quantification and sample throughput by mixing multiple samples and standards, each tagged with different sets of CS probes. |
Analysis of Macromolecules
Macromolecules such as proteins, nucleic acids and complex carbohydrates are major components of biomass and are products of anabolic metabolism. As such they are principal sinks and sources of carbon, nitrogen (and phosphorus) in cells and tissues. SIRM-based studies need to account for the isotopic enrichment in these components. One approach is to isolate the bulk macromolecules, and digest them to their component subunits (i.e. amino acids, nucleotides and monosaccharides) and analyze them by mass spectrometry and NMR . Many traditional methods of hydrolysis are time consuming and lead to significant degradation, thus biasing the results. We have been developing methods for rapid hydrolysis of proteins and complex carbohydrates such as glycogen that preserve the integrity of the subunits, so that isotopologue distributions and net isotope incorporation can be determined reliably.References
- Fan TW-M. Sample Preparation for Metabolomics Investigation. In: Fan TW-M, Lane AN, Higashi RM, eds. The Handbook of Metabolomics: Pathway and Flux Analysis, Methods in Pharmacology and Toxicology DOI 101007/978-1-61779-618-0_11. New York: Springer Science; 2012:7-27.
- SS Gori, P. Lorkiewicz, DS Ehringer, AC Belshoff, RM Higashi, TW-M Fan and MH Nantz (2014) Profiling Thiol Metabolites and Quantification of Cellular Glutathione using FT-ICR-MS Spectrometry. Analytical & Bioanalytical Chemistry 406, 4371-9
-
Fan, T. W-M.,Tan, J., McKinney, M.M. and Lane, A.N. (2011). Stable Isotope Resolved Metabolomics Analysis of Ribonucleotide and RNA Metabolism in Human Lung Cancer Cells. Metabolomics 8, 517-527
- Lane, A.N., Arumugam, S., Lorkiewicz P.K., Higashi, R.M., Laulhe, S., Nantz, M.H., Moseley, H.N.B., Fan, T. W-M. (2015) Chemoselective detection of carbonyl compounds in metabolite mixtures by NMR. Mag Res Chem. 53, 337-343
-
Lane, A.N., Fan, T. W-M., Yan, J. (2015) 13C Tracer studies in mouse tumor xenografts. Bio-protocol 5(22): e1650
-
Fan, T. W-M.,Lane, A.N., Higashi, R.M. (2016) Stable Isotope Resolved Metabolomics studies in ex vivo tissue slices. Bio-protocol 6, e1730
-
Yang, Y., Fan, W. W-M., Lane, A.N. & Higashi, R.M. (2017) ChloroformateDerivatization for Tracing the Fate of Amino Acids in Cells by Multiple Stable Isotope Resolved Metabolomics (mSIRM). Anal. Chim. Acta 976, 63-73
- Deng, P., Higashi, R.M., Lane, A.N., Bruntz, R.C., Sun, R.C., Nantz, M.H., Fan, T. W-M. (2018) Quantification and Identification of Carbonyls Using Chemoselective Tagging and NanoESI FT-MS. Analyst 143, 311-322
-
Fan, T. W-M., El-Amouri, S.S., Macedo, J.K.A., Wang, Q.J. and Lane, A.N. (2018) Mapping Metabolic Networks in 3D Spheroids Using Stable Isotope-Resolved Metabolomics. Metabolites 8, 40
 Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors. Ideas, requests, problems regarding Moseley Bioinformatics Lab? Send feedback