You are here: Moseley Bioinformatics Lab>Main Web>ResearchInterests (06 Jul 2024, HunterMoseley)
Moseley Laboratory Research Interests
Broad Theme:
Develop computational methods/models/tools for analyzing, integrating, and interpreting many types of biological and biophysical data that enable new understanding of biological systems and related disease processes.
Our approach involves:
- Leveraging relevant information from large public scientific repositories and knowledgebases.
- Developing appropriate methods to analyze specific types of biological data.
- Creating new models that facilitate the integration of diverse types of biological data.
- Implementing system-wide analyses that integrate omics-level datasets.
Specific Interests:
Systems Biochemical Tools for Large-Scale Stable-Isotope Resolved Metabolomics (SIRM) Applications
Our lab provides bioinformatics and systems biology expertise for the analysis and interpretation of SIRM experiments. Our goal is to develop a combination of bioinformatic, biostatistical, and systems biochemical tools implemented in an integrated data analysis pipeline that will allow broad application of SIRM from the discovery of specific metabolic phenotypes representing biological and disease states of interest to a mechanism-based understanding of a wide range of specific human disease processes with particular metabolic phenotypes. Our new tools are already providing novel metabolic pathway-specific analyses of complex SIRM datasets. For example, we have used a moiety model analysis of SIRM mass spectrometer data to quantitate the relative importance of specific metabolic pathways in the biosynthesis of UDP-GlcNAc in prostate cancer cell culture. Subsequent analyses determined which pathways were impacted by potential cancer therapeutics. As we implement a complete SIRM-based data analysis pipeline, our ultimate goal is to integrate metabolomics datasets with other major omics datasets including epigenomics, genomics, transcriptomics, and proteomics datasets in full systems biochemical analyses that can determine which gene-regulatory, signaling, and metabolic pathways are mechanistically involved in specific human diseases.
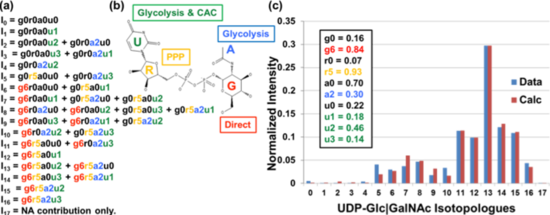 |
Figure 1: (a) Chemical substructure model representing the possible number of 13C incorporation from 13C6-Glc tracer into UDP-GlcNAc, accounting for the observed FT-ICR-MS isotopologue peaks. (b) Structure of UDP-GlcNAc annotated by its chemical substructures and their biosynthetic pathways from 13C6-Glc, as in Fig. 2. U = uracil, R = ribose, A = acetyl, G=glucose. NAc-Glucose utilizes Gln as the nitrogen donor. (c) Fit of optimized chemical substructure model parameters to FT-ICR-MS isotopologue data of UDP-GlcNAc extracted from a LN3 prostate cancer cell culture after 48 hours of growth in 13C6-Glc. |
- Hunter N.B. Moseley. "Correcting for the Effects of Natural Abundance in Stable Isotope Resolved Metabolomics Experiments Involving Ultra-High Resolution Mass Spectrometry." BMC Bioinformatics 11, 139-144 (2010).
- Hunter N.B. Moseley, Andrew N. Lane, Alex C. Belshoff, Richard M. Higashi, and Teresa W-M. Fan. "A novel method for deconvoluting metabolic subunits from mass isotopologues in stable isotope resolved metabolomic experiments under non steady-state conditions: application to the biosynthesis of UDP-GlcNAc" BMC Biology, 9, 37 (2011).
- Hunter N.B. Moseley. "Error Analysis and Propagation in Metabolomics Data Analysis" Comp Struct Biotech J, 4, e201301006 (2013).
- Joshua M. Mitchell, Teresa, W-M. Fan, Andrew N. Lane, Hunter N.B. Moseley. "Development and In silico Evaluation of Large-Scale Metabolite Identification Methods using Functional Group Detection for Metabolomics" Frontiers in Genetics - Systems Biology 5, 237 (2014).
- Joshua M. Mitchell, Robert M. Flight, QingJun Wang, Richard M Higashi, Teresa W-M Fan, Andrew N. Lane, and Hunter N.B. Moseley. "New Methods to Identify High Peak Density Artifacts in Fourier Transform Mass Spectra and mitigate their Effects on High-Throughput Metabolomics Data Analysis" Metabolomics 14, 125 (2018).
- Huan Jin and Hunter N.B. Moseley. "Moiety Modeling Framework for Deriving Moiety Abundances from Mass Spectrometry Measured Isotopologues" BMC Bioinformatics 20, 524 (2019).
- Joshua M. Mitchell, Robert M. Flight, and Hunter N.B. Moseley. "Small Molecule Isotope Resolved Formula Enumeration: a Methodology for Assigning Isotopologues and Metabolites in Fourier Transform Mass Spectra" Analytical Chemistry 91, 8933 (2019).
- Huan Jin and Hunter N.B. Moseley. "Robust Moiety Model Selection Using Mass Spectrometry Measured Isotopologues" Metabolites 10, 118 (2020).
- Huan Jin, Joshua M. Mitchell, and Hunter N.B. Moseley. "Atom Identifiers Generated by a Neighborhood-Specific Graph Coloring Method Enable Compound Harmonization Across Metabolic Databases" Metabolites 10, 368 (2020).
- Huan Jin and Hunter N.B. Moseley. "Hierarchical Harmonization of Atom-Resolved Metabolic Reactions Across Metabolic Databases" Metabolites 11, 431 (2021).
- Robert M. Flight, Joshua M. Mitchell, and Hunter N.B. Moseley. "Scan-Centric, Frequency-Based Method for Characterizing Peaks from Direct Injection Fourier transform Mass Spectrometry Experiments" Metabolites 12, 515 (2022).
- Huan Jin and Hunter N.B. Moseley. "md_harmonize: a Python package for atom-level harmonization of public metabolic databases" Metabolites 13, 1199 (2023).
Metabolome Mining
Metabolome mining uses known and/or predicted metabolite annotations to derive metabolic information that is interpretable in a biological or biomedical context. Often metabolome mining methods aggregate unassigned (Level 5) or partially assigned (lower than Level 1 validated assignments) metabolite features along with associated chemical or biochemical annotations in order to improve statistical power. Our lab is pioneering the development of metabolome mining methods that utilize predicted metabolite annotations to derive meaningful information from metabolomics datasets.
- Joshua M. Mitchell, Robert M. Flight, and Hunter N.B. Moseley. "Small Molecule Isotope Resolved Formula Enumeration: a Methodology for Assigning Isotopologues and Metabolites in Fourier Transform Mass Spectra" Analytical Chemistry 91, 8933 (2019).
- Joshua M. Mitchell, Robert M. Flight, and Hunter N.B. Moseley. "Deriving Accurate Lipid Classification based on Molecular Formula" Metabolites 10, 122 (2020).
- Joshua M. Mitchell, Robert M. Flight, and Hunter N.B. Moseley. "Untargeted lipidomics of non-small cell lung carcinoma demonstrates differentially abundant lipid classes in cancer vs non-cancer tissue" Metabolites 11, 740 (2021).
- Erik D. Huckvale, Christian D. Powell, Huan Jin, and Hunter N.B. Moseley. "Benchmark dataset for training machine learning models to predict the pathway involvement of metabolites" Metabolites 13, 1120 (2023).
- Hunter N.B. Moseley. "In the AI science boom, beware: your results are only as good as your data -- Machine-learning systems are voracious data consumers -- but trustworthy results require more vetting both before and after publication" Nature Feb 1 (2024).
- Erik D. Huckvale and Hunter N.B. Moseley. "A cautionary tale about properly vetting datasets used in supervised learning predicting metabolic pathway involvement" PLOS One 19, e0299583 (2024).
- Erik D. Huckvale and Hunter N.B. Moseley. "Predicting The Pathway Involvement Of Metabolites Based on Combined Metabolite and Pathway Features" Metabolites 14, 266 (2024).
FAIR Data Sharing and Open Science
The FAIR (Findable, Accessible, Interoperable, and Reusable) Guiding Principles of Data Stewardship are a major part of Open Science, with the goal to make all research data, products, and knowledge openly accessible by anyone, thereby promoting collaborative research efforts. Our lab has developed a variety of open-source tools that promote the FAIRness of specific data repositories and knowledgebases. For example, we have developed the open-source mwtab Python library and package for FAIRer access to Metabolomics Workbench as well as the Metabolomics Workbench Validation website that provides weekly evaluations of all datasets made available in the repository with respect to consistency and conformity to repository deposition standards. Also, we have developed the open-source nmrstarlib Python library for FAIRer access to the Biological Magnetic Resonance Data Bank and the open-source kegg_pull Python package for FAIRer access to the Kyoto Encyclopedia of Gene and Genomes (KEGG). Moreover, we have extended and developed new data deposition standards when such standards were lacking or missing. For example, we developed the draft Minimum Information About Geospatial Information System (MIAGIS) standard for facilitating public deposition of geospatial information system (GIS) datasets as well as the open-source miagis Python package that facilitates generation of the MIAGIS deposition format. Recently, we developed the open-source MESSES Python package for comprehensive (meta)data capture, validation, and conversion into mwTab deposition format.
All Python packages are available through the Python Package Index (PyPI) and GitHub with extensive end-user documentation. Similarly, all R packages are available via GitHub and CRAN or Bioconductor with comprehensive end-user documentation vignettes. All GitHub repositories are organized and managed under the Moseley Bioinformatics and Systems Biology Lab organizational account: https://github.com/MoseleyBioinformaticsLab
- Yasset Perez-Riverol, Laurent Gatto,Rui Wang, Timo Sachsenberg, Julian Uszkoreit, Felipe da Veiga Leprevost, Christian Fufezan,Tobias Ternent, Stephen J. Eglen, Daniel S. Katz, Tom J. Pollard, Alexander Konovalov, Robert M. Flight, Kai Blin, Juan Antonio Vizcaíno. "Ten Simple Rules for Taking Advantage of Git and GitHub" PLOS One 12, e1004947 (2016).
- Andrey Smelter, Morgan Astra, Hunter N.B. Moseley. "A Fast and Efficient Python Library for Interfacing with the Biological Magnetic Resonance Data Bank" BMC Bioinformatics 18, 175 (2017).
- Andrey Smelter and Hunter N.B. Moseley. "A Python library for FAIRer access and deposition to the Metabolomics Workbench Data Repository" Metabolomics 14, 64 (2018).
- Sen Yao and Hunter N.B. Moseley. "A chemical interpretation of protein electron density maps in the worldwide protein data bank" PLOS One 15, e0236894 (2020).
- Christian D. Powell and Hunter N.B. Moseley. "The mwtab Python library for RESTful Access and Enhanced Quality Control, Deposition, and Curation of the Metabolomics Workbench Data Repository" Metabolites 11, 163 (2021).
- P. Travis Thompson, Christian D. Powell, and Hunter N.B. Moseley. "Academic Tracker: Software for Tracking and Reporting Publications Associated with Authors and Grants" PLOS One 17, e0277834 (2022).
- Erik Huckvale and Hunter N.B. Moseley. "kegg_pull: a Software Package for the RESTful Access and Pulling from The Kyoto Encyclopedia of Gene and Genomes" BMC Bioinformatics 24, 78 (2023).
- P. Travis Thompson, Sweta Ojha, Christian D. Powell, Kelly G. Pennell, and Hunter N.B. Moseley. "A proposed FAIR approach for disseminating geospatial information system maps" Scientific Data 10, 389 (2023).
- Rance Nault, Matthew C. Cave, Gabriele Ludewig, Hunter N.B. Moseley, Kelly G. Pennell, and Tim Zacharewski. "A case for accelerating standards to achieve the FAIR principles of environmental health research experimental data" Environmental Health Perspectives 131, 6 (2023).
- Christian D. Powell and Hunter N.B. Moseley. "The Metabolomics Workbench File Status Website: A Metadata Repository Promoting FAIR Principles of Metabolomics Data" BMC Bioinformatics 24, 299 (2023).
- P. Travis Thompson and Hunter N.B. Moseley. "MESSES: Software for Transforming Messy Research Datasets into Clean Submissions to Metabolomics Workbench for Public Sharing" Metabolites 13, 842 (2023).
- Hunter N.B. Moseley, Philippe Rocca-Serra, Reza M. Salek, Masanori Arita, and Emma L. Schymanski. "InChI Isotopologue and Isotopomer Specifications" J Cheminformatics 16, 54 (2024).
Improved Utilization and Curation of the Gene Ontology with Interaction Network Integration
The Gene Ontology (GO) is the largest and best curated ontology in the OBO Foundry and is used extensively to precisely describe the functions, locations, and processes of gene(-product)s through specific annotations stored across many knowledgebases. But there is a fundamental problem with a lack of tools that organize ontology terms into usable domain-specific concepts that biomedical researchers can easily interpret, leverage within statistically rigorous analyses, and integrate with other types of information. Therefore, we have developed the GO Categorization Suite (GOcats), which streamlines the slicing of GO into custom, biologically-meaningful subgraphs representing emergent concepts in GO. GOcats uses a list of user-defined keywords or GO terms that describe a concept, the structure of GO, and relationship properties to automatically generate a subgraph of child terms and a mapping of these child terms to their respective concept-defining term. GOcats enables the utilization of additional GO relationship types in a manner that preserves proper scoping and scaling. Furthermore, we have demonstrated improvements in statistical power via the use of GOcats in annotation enrichment analyses performed by categoryCompare. We have also integrated GOcats driven annotation enrichment analysis with principal component analysis and molecular interaction network analysis (see Figure). Moreover, we have collaborated in the development of advanced curation tools that can help detect missing and erroneous relationships in GO, which are needed due to GO’s size (over 40,000 terms) and rate of growth.
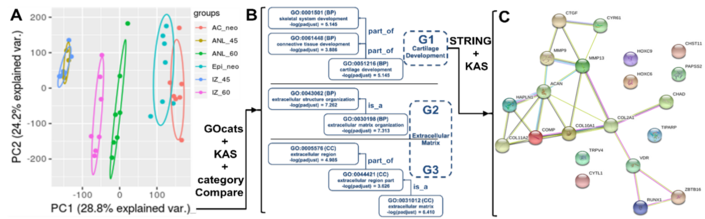 |
Figure 2. A) PCA plot of equine RNAseq datasets. B) Organized groups of enriched GO-terms for PC1. C) STRING interactions between high PC1 loading gene(-product)s annotated with group G1 GO terms (cartilage development). |
- Rashmie Abeysinghe, Eugene W. Hinderer III, Hunter N.B. Moseley, and Licong Cui. "Auditing Subtype Inconsistencies among Gene Ontology Concepts" The 2nd International Workshop on Semantics-Powered Data Analytics (SEPDA 2017) -- Bioinformatics and Biomedicine (BIBM), 2017 IEEE International Conference 1242-1245 (2017).
- Rashmie Abeysinghe, Fengbo Zheng, Eugene W. Hinderer III, Hunter N.B. Moseley, and Licong Cui. "A Lexical Approach to Identifying Subtype Inconsistencies in Biomedical Terminologies" Quality Assurance of Biological and Biomedical Ontologies and Terminologies Workshop -- Bioinformatics and Biomedicine (BIBM), 2018 IEEE International Conference pp. 1982-1989 (2018).
- Eugene W. Hinderer III, Robert M. Flight, Rashmi Dubey, James N. MacLeod, and Hunter N.B. Moseley. "Advances in Gene Ontology Utilization Improve Statistical Power of Annotation Enrichment" PLOS One 14, e0220728 (2019).
- Eugene W. Hinderer III and Hunter N.B. Moseley. "GOcats: A tool for categorizing Gene Ontology into subgraphs of user-defined concepts" PLOS One 15, e0233311 (2020).
- Rashmie Abeysinghe, Eugene W. Hinderer III, Hunter N.B. Moseley, and Licong Cui. "SSIF:Subsumption-based Sub-term Inference Framework to Audit Gene Ontology" Bioinformatics 36, 3207 (2020).
Older Interests:
Interaction Network-centric Cancer Mutational Pattern Analyses
Lung cancer is the leading cause of cancer death worldwide, with 160,000 deaths in the US annually. The state of Kentucky ranks highest in lung cancer incidence and mortality, with the Central Appalachian region of Kentucky (AppKY) ranking the highest of the highest. Squamous cell carcinoma (SQCC) of the lung from AppKY has uniquely high mutation rates in PCMTD1 and IDH1 genes in comparison to The Cancer Genome Atlas (TCGA), suggesting that pathways including these genes are likely important for cancer development in this population. Therefore, we have developed analyses for placing these genes within molecular interaction networks constructed from known protein-protein interactions and gene-products with related function. In this application, we have found mutually exclusive mutational patterns between PCMTD1 and related histone methylases and between IDH1 and related histone demethylases, suggesting that mutations in these pathways directing histone methylation and demethylation are important in SQCC cancer development and may be related to AppKY-specific environmental factors.
Structural Bioinformatics of Metalloproteins
| Structural bioinformatics of metalloproteins has historically been hampered by significant numbers of aberrant coordination geometries that prevented systematic classification. My lab has developed combined functional and structural analyses of metalloproteins that have identified aberrant clusters of coordination geometries (CG) of metal ion ligation in the top 5 most abundant metalloproteins. Most of these aberrant CGs are due to multidentate ligands that create compressed ligand-metal-ligand angles below 60°. These angles cause serious deviations from canonical CG models and greatly hamper the ability to characterize metalloproteins both structurally and functionally. Our methods detect coordinating ligands without expectations based on canonical CGs and in a statistically robust manner, producing estimated false positive and false negative rates of ~0.11% and ~1.2%, respectively. Also, our improved analyses of bond-length distributions have revealed bond-length modes specific to chemical functional groups involved in multidentation. By recognizing aberrant CGs in our clustering analyses, high correlations above 0.9 are achieved between structural and functional descriptions of metal ion coordination. This work has been impactful to the field by highlighting the unexpected presence of significant numbers of non-canonical CGs and in characterizing their structural, functional, and chemical characteristics. Our publications made the cover of the May 2017 issue of Proteins. |  |
- Sen Yao, Robert M. Flight, Eric C. Rouchka, and Hunter N.B. Moseley. "A less biased analysis of metalloproteins reveals novel zinc coordination geometries" Proteins: Structure, Function, and Bioinformatics 83, 1470 (2015).
- Sen Yao, Robert M. Flight, Eric C. Rouchka, Hunter N.B. Moseley. "Aberrant coordination geometries discovered in the most abundant metalloproteins" Proteins: Structure, Function, and Bioinformatics 85, 885 (2017).
- Sen Yao, Robert M. Flight, Eric C. Rouchka, Hunter N.B. Moseley. "Perspectives and Expectations in Structural Bioinformatics of Metalloproteins" Proteins: Structure, Function, and Bioinformatics 85, 938 (2017).
- Sen Yao and Hunter N.B. Moseley. "Finding high-quality metal ion-centric regions across the worldwide Protein Data Bank" Molecules 24, 3179 (2019).
- Sen Yao and Hunter N.B. Moseley. "A chemical interpretation of protein electron density maps in the worldwide protein data bank" PLOS One 15, e0236894 (2020).
- Alan Luo and Hunter N.B. Moseley. "Electron Density Discrepancy Analysis of Energy Metabolism Coenzymes" International Journal of Bioengineering and Life Sciences 17, 307 (2023).
Automated Analysis Tools for Magic Angle Spinning Solid State NMR Protein Resonance Data
Membrane proteins are essential for many biological functions. They comprise roughly one third of all sequenced genomes, and represent 70% of all current drug targets. However, fewer than 1500 of the ~100,000 protein structure entries in the worldwide Protein Data Bank (PDB) involve integral membrane proteins as of June 2009. This is because they are difficult to crystallize for x-ray crystallographic studies and difficult to solubilize for solution nuclear magnetic resonance (NMR) studies. Magic-angle spinning solid-state NMR (MAS SSNMR) represents a fast developing experimental method that has great potential to provide structural and dynamics information of membrane proteins without the sample limitations of other techniques. We are developing automated analysis tools that will aid in the analysis of SSNMR data and specifically tailored for SSNMR data from membrane protein samples. Specifically our lab is focusing on developing and testing algorithms that will automate all analysis steps from raw SSNMR spectral data to protein resonance assignments for uniformly 13C/15N-labeled membrane proteins. This development will provide necessary analysis tools for expansion of MAS SSNMR and its application to membrane proteins into the broader biological community.
Topic revision: r41 - 06 Jul 2024, HunterMoseley
 Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors.
Copyright © by the contributing authors. All material on this collaboration platform is the property of the contributing authors. Ideas, requests, problems regarding Moseley Bioinformatics Lab? Send feedback
